Lineage for d1yjnf1 (1yjn F:1-119)
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.79: Bacillus chorismate mutase-like [55297] (9 superfamilies)
core: beta-alpha-beta-alpha-beta(2); mixed beta-sheet: order: 1423, strand 4 is antiparallel to the rest
Superfamily d.79.3: L30e-like [55315] (4 families) 

Family d.79.3.1: L30e/L7ae ribosomal proteins [55316] (4 proteins) 
Protein Ribosomal protein L7ae [55319] (7 species) 
Species Haloarcula marismortui [TaxId:2238] [55320] (58 PDB entries)
Uniprot P12743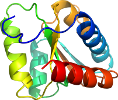
Domain d1yjnf1: 1yjn F:1-119 [123439]
Other proteins in same PDB: d1yjn11, d1yjn21, d1yjn31, d1yjna1, d1yjna2, d1yjnb1, d1yjnc1, d1yjnd1, d1yjne1, d1yjne2, d1yjng1, d1yjnh1, d1yjni1, d1yjnj1, d1yjnk1, d1yjnl1, d1yjnm1, d1yjnn1, d1yjno1, d1yjnp1, d1yjnq1, d1yjnr1, d1yjns1, d1yjnt1, d1yjnu1, d1yjnv1, d1yjnw1, d1yjnx1, d1yjny1, d1yjnz1
automatically matched to d1s72f_
complexed with cd, cl, cly, k, mg, na; mutant
Details for d1yjnf1
PDB Entry: 1yjn (more details), 3 Å
PDB Description: crystal structure of clindamycin bound to the g2099a mutant 50s ribosomal subunit of haloarcula marismortui
PDB Compounds: (F:) 50S ribosomal protein L7AeSCOPe Domain Sequences for d1yjnf1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1yjnf1 d.79.3.1 (F:1-119) Ribosomal protein L7ae {Haloarcula marismortui [TaxId: 2238]}
pvyvdfdvpadleddalealevardtgavkkgtnettksiergsaelvfvaedvqpeeiv
mhipeladekgvpfifveqqddlghaaglevgsaaaavtdageadadvediadkveelr
SCOPe Domain Coordinates for d1yjnf1:
Click to download the PDB-style file with coordinates for d1yjnf1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1yjnf1:
- d1yjnf1 first appeared in SCOP 1.73
- d1yjnf1 appears in SCOPe 2.06
- d1yjnf1 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1yjn11, d1yjn21, d1yjn31, d1yjna1, d1yjna2, d1yjnb1, d1yjnc1, d1yjnd1, d1yjne1, d1yjne2, d1yjng1, d1yjnh1, d1yjni1, d1yjnj1, d1yjnk1, d1yjnl1, d1yjnm1, d1yjnn1, d1yjno1, d1yjnp1, d1yjnq1, d1yjnr1, d1yjns1, d1yjnt1, d1yjnu1, d1yjnv1, d1yjnw1, d1yjnx1, d1yjny1, d1yjnz1 |