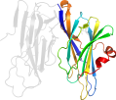Lineage for d1yjka1 (1yjk A:50-198)
- Root: SCOPe 2.06

Class b: All beta proteins [48724] (177 folds) 
Fold b.121: Nucleoplasmin-like/VP (viral coat and capsid proteins) [88632] (7 superfamilies)
sandwich; 8 strands in 2 sheets; jelly-roll; some members can have additional 1-2 strands
characteristic interaction between the domains of this fold allows the formation of five-fold and pseudo six-fold assemblies
Superfamily b.121.1: PHM/PNGase F [49742] (2 families) 
members of this superfamily bind peptide substrates
duplication: consists of two domains of this fold packed together like the adjacent nucleoplasmin subunits
Family b.121.1.2: Peptidylglycine alpha-hydroxylating monooxygenase, PHM [49746] (1 protein) 
Protein Peptidylglycine alpha-hydroxylating monooxygenase, PHM [63402] (1 species) 
Species Norway rat (Rattus norvegicus) [TaxId:10116] [49748] (17 PDB entries) 
Domain d1yjka1: 1yjk A:50-198 [123423]
automated match to d3miha1
complexed with cu, gol
Details for d1yjka1
PDB Entry: 1yjk (more details), 2 Å
PDB Description: Reduced Peptidylglycine Alpha-Hydroxylating Monooxygenase (PHM) in a New Crystal Form
PDB Compounds: (A:) Peptidyl-glycine alpha-amidating monooxygenaseSCOPe Domain Sequences for d1yjka1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1yjka1 b.121.1.2 (A:50-198) Peptidylglycine alpha-hydroxylating monooxygenase, PHM {Norway rat (Rattus norvegicus) [TaxId: 10116]}
tigpvtpldasdfaldirmpgvtpkesdtyfcmsmrlpvdeeafvidfkprasmdtvhhm
llfgcnmpsstgsywfcdegtctdkanilyawarnapptrlpkgvgfrvggetgskyfvl
qvhygdisafrdnhkdcsgvsvhltrvpq
SCOPe Domain Coordinates for d1yjka1:
Click to download the PDB-style file with coordinates for d1yjka1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1yjka1:
- d1yjka1 first appeared in SCOP 1.73
- d1yjka1 appears in SCOPe 2.05
- d1yjka1 appears in SCOPe 2.07
- d1yjka1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D