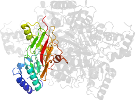
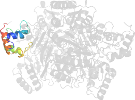Lineage for d1wveb1 (1wve B:243-521)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.58: Ferredoxin-like [54861] (59 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.32: FAD-linked oxidases, C-terminal domain [55103] (4 families) 
duplication: contains two subdomains of this fold
Family d.58.32.1: Vanillyl-alcohol oxidase-like [55104] (2 proteins) 
Protein Flavoprotein subunit of p-cresol methylhydroxylase [55107] (1 species)
the other subunit is a short-chain cytochrome c
Species Pseudomonas putida [TaxId:303] [55108] (4 PDB entries) 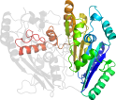
Domain d1wveb1: 1wve B:243-521 [121333]
Other proteins in same PDB: d1wvea2, d1wveb2, d1wvec1, d1wved1
automatically matched to d1diia1
complexed with acy, cl, fad, hem, trs
Details for d1wveb1
PDB Entry: 1wve (more details), 1.85 Å
PDB Description: p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit
PDB Compounds: (B:) 4-cresol dehydrogenase [hydroxylating] flavoprotein subunitSCOP Domain Sequences for d1wveb1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1wveb1 d.58.32.1 (B:243-521) Flavoprotein subunit of p-cresol methylhydroxylase {Pseudomonas putida [TaxId: 303]}
pvfkpfevifedeadiveivdalrplrmsntipnsvviastlweagsahltraqyttepg
htpdsvikqmqkdtgmgawnlyaalygtqeqvdvnwkivtdvfkklgkgrivtqeeagdt
qpfkyraqlmsgvpnlqefglynwrggggsmwfapvseargseckkqaamakrvlhkygl
dyvaefivaprdmhhvidvlydrtnpeetkradacfnelldefekegyavyrvntrfqdr
vaqsygpvkrklehaikravdpnnilapgrsgidlnndf
SCOP Domain Coordinates for d1wveb1:
Click to download the PDB-style file with coordinates for d1wveb1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1wveb1:
- d1wveb1 first appeared in SCOP 1.73
- d1wveb1 appears in SCOPe 2.01
- d1wveb1 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1wvea1, d1wvea2, d1wvec1, d1wved1 |