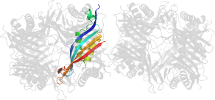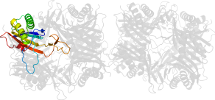Lineage for d1vayg1 (1vay G:142-297)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.96: T-fold [55619] (2 superfamilies)
beta(2)-alpha(2)-beta(2); 2 layers: alpha/beta; antiparallel sheet 1234
tunnel-shaped: its known members form wide oligomeric barrels different sizes
Superfamily d.96.1: Tetrahydrobiopterin biosynthesis enzymes-like [55620] (4 families) 
bind purine or pterin in topologically similar sites between subunits
Family d.96.1.4: Urate oxidase (uricase) [55633] (1 protein) 
Protein Urate oxidase (uricase) [55634] (3 species)
duplication: one subunit consists of two domains of this fold; beta-sheets of two subunits form a barrel, closed: n=16, S=16
Species Arthrobacter globiformis [TaxId:1665] [143630] (6 PDB entries) 
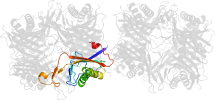
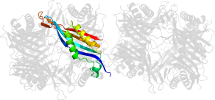
Domain d1vayg1: 1vay G:142-297 [119938]
automatically matched to 1VAX A:142-297
complexed with aza
Details for d1vayg1
PDB Entry: 1vay (more details), 2.24 Å
PDB Description: Crystal Structure of Uricase from Arthrobacter globiformis with inhibitor 8-azaxanthine
PDB Compounds: (G:) Uric acid oxidaseSCOP Domain Sequences for d1vayg1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1vayg1 d.96.1.4 (G:142-297) Urate oxidase (uricase) {Arthrobacter globiformis [TaxId: 1665]}
seqaivagiegltvlkstgsefhgfprdkyttlqettdrilatdvsarwryntvevdfda
vyasvrglllkafaethslalqqtmyemgraviethpeideikmslpnkhhflvdlqpfg
qdnpnevfyaadrpyglieatiqregsradhpiwsn
SCOP Domain Coordinates for d1vayg1:
Click to download the PDB-style file with coordinates for d1vayg1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1vayg1:
- d1vayg1 first appeared in SCOP 1.73
- d1vayg1 appears in SCOPe 2.01
- d1vayg1 appears in the current release, SCOPe 2.08
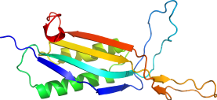
 View in 3D
View in 3D