Lineage for d1y9ma1 (1y9m A:373-536)
- Root: SCOP 1.71

Class b: All beta proteins [48724] (149 folds) 
Fold b.29: Concanavalin A-like lectins/glucanases [49898] (1 superfamily)
sandwich; 12-14 strands in 2 sheets; complex topology
Superfamily b.29.1: Concanavalin A-like lectins/glucanases [49899] (21 families) 

Family b.29.1.19: Glycosyl hydrolases family 32 C-terminal domain [101652] (2 proteins)
Pfam 08244
flat sheet beta-sandwich lacking the characteristic beta-bulge in the C-terminal strand
Protein Exo-inulinase [117137] (1 species) 
Species Aspergillus awamori [TaxId:105351] [117138] (3 PDB entries) 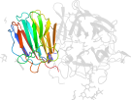
Domain d1y9ma1: 1y9m A:373-536 [116590]
Other proteins in same PDB: d1y9ma2
complexed with gol, man, nag
Details for d1y9ma1
PDB Entry: 1y9m (more details), 1.89 Å
PDB Description: crystal structure of exo-inulinase from aspergillus awamori in spacegroup p212121
SCOP Domain Sequences for d1y9ma1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1y9ma1 b.29.1.19 (A:373-536) Exo-inulinase {Aspergillus awamori}
eawssisnkrpiysrtfktlsegstnttttgetfkvdlsfsakskastfaialrasanft
eqtlvgydfakqqifldrthsgdvsfdetfasvyhgpltpdstgvvklsifvdrssvevf
ggqgettltaqifpssdavharlastggttedvradiykiastw
SCOP Domain Coordinates for d1y9ma1:
Click to download the PDB-style file with coordinates for d1y9ma1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1y9ma1:
- d1y9ma1 is new in SCOP 1.71
- d1y9ma1 appears in SCOP 1.73
- d1y9ma1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D