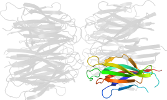
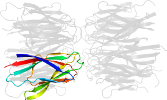
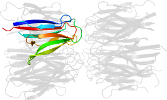
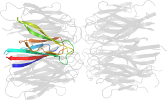
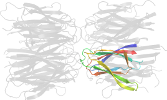
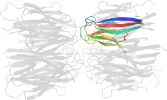
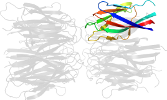
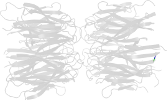
Lineage for d1xe0f1 (1xe0 F:16-122)
- Root: SCOPe 2.07

Class b: All beta proteins [48724] (178 folds) 
Fold b.121: Nucleoplasmin-like/VP (viral coat and capsid proteins) [88632] (7 superfamilies)
sandwich; 8 strands in 2 sheets; jelly-roll; some members can have additional 1-2 strands
characteristic interaction between the domains of this fold allows the formation of five-fold and pseudo six-fold assemblies
Superfamily b.121.3: Nucleoplasmin-like core domain [69203] (2 families) 
oligomerizes into a pentameric ring structure
Family b.121.3.1: Nucleoplasmin-like core domain [69204] (4 proteins)
automatically mapped to Pfam PF03066
Protein Nucleophosmin (NO38) [117086] (1 species) 
Species African clawed frog (Xenopus laevis) [TaxId:8355] [117087] (2 PDB entries)
Uniprot P07222 16-122
Domain d1xe0f1: 1xe0 F:16-122 [115202]
Other proteins in same PDB: d1xe0a2, d1xe0b2, d1xe0d2, d1xe0e2, d1xe0f2, d1xe0g2, d1xe0h2, d1xe0i2, d1xe0j2
Details for d1xe0f1
PDB Entry: 1xe0 (more details), 1.7 Å
PDB Description: The structure and function of Xenopus NO38-core, a histone binding chaperone in the nucleolus
PDB Compounds: (F:) NucleophosminSCOPe Domain Sequences for d1xe0f1:
Sequence, based on SEQRES records: (download)
>d1xe0f1 b.121.3.1 (F:16-122) Nucleophosmin (NO38) {African clawed frog (Xenopus laevis) [TaxId: 8355]}
qnflfgcelkadkkeysfkveddenehqlslrtvslgasakdelhvveaeginyegktik
ialaslkpsvqptvslggfeitppvilrlksgsgpvyvsgqhlvale
Sequence, based on observed residues (ATOM records): (download)
>d1xe0f1 b.121.3.1 (F:16-122) Nucleophosmin (NO38) {African clawed frog (Xenopus laevis) [TaxId: 8355]}
qnflfgcelkadkkeysfkvedenehqlslrtvslgasakdelhvveaeginyegktiki
alaslkpsvqptvslggfeitppvilrlksgsgpvyvsgqhlvale
SCOPe Domain Coordinates for d1xe0f1:
Click to download the PDB-style file with coordinates for d1xe0f1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1xe0f1:
- d1xe0f1 first appeared in SCOP 1.71, called d1xe0f_
- d1xe0f1 appears in SCOPe 2.06
- d1xe0f1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D