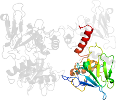Lineage for d1x6ma1 (1x6m A:4-196)
- Root: SCOPe 2.06

Class b: All beta proteins [48724] (177 folds) 
Fold b.88: Mss4-like [51315] (2 superfamilies)
complex fold made of several coiled beta-sheets
Superfamily b.88.1: Mss4-like [51316] (5 families) 
duplication: tandem repeat of two similar structural motifs
Family b.88.1.4: Glutathione-dependent formaldehyde-activating enzyme, Gfa [117338] (1 protein)
Pfam PF04828; DUF636
Protein Glutathione-dependent formaldehyde-activating enzyme, Gfa [117339] (1 species) 
Species Paracoccus denitrificans [TaxId:266] [117340] (2 PDB entries)
Uniprot Q51669
Domain d1x6ma1: 1x6m A:4-196 [114920]
Other proteins in same PDB: d1x6ma2, d1x6mb2, d1x6mc2, d1x6md2
complexed with gol, so4, zn
Details for d1x6ma1
PDB Entry: 1x6m (more details), 2.35 Å
PDB Description: crystal structure of the glutathione-dependent formaldehyde-activating enzyme (gfa)
PDB Compounds: (A:) Glutathione-dependent formaldehyde-activating enzymeSCOPe Domain Sequences for d1x6ma1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1x6ma1 b.88.1.4 (A:4-196) Glutathione-dependent formaldehyde-activating enzyme, Gfa {Paracoccus denitrificans [TaxId: 266]}
vdtsgvkihpavdngikpaqpgfaggtlhckcstnpvrvavraqtahnhvcgctkcwkpe
gaifsqvavvgrdalevlegaekleivnaeapiqrhrcrdcgvhmygrienrdhpfygld
fvhtelsdedgwsapefaafvssiiesgvdpsrmeairarlrelglepydalspplmdai
athiakrsgalaa
SCOPe Domain Coordinates for d1x6ma1:
Click to download the PDB-style file with coordinates for d1x6ma1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1x6ma1:
- d1x6ma1 first appeared in SCOP 1.71, called d1x6ma_
- d1x6ma1 was called d1x6ma_ in SCOPe 2.05
- d1x6ma1 appears in SCOPe 2.07
- d1x6ma1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D