Lineage for d1vmia_ (1vmi A:)
- Root: SCOP 1.73

Class c: Alpha and beta proteins (a/b) [51349] (141 folds) 
Fold c.77: Isocitrate/Isopropylmalate dehydrogenase-like [53658] (1 superfamily)
consists of two intertwined (sub)domains related by pseudo dyad; duplication
3 layers: a/b/a; single mixed beta-sheet of 10 strands, order 213A945867 (A=10); strands from 5 to 9 are antiparallel to the rest
Superfamily c.77.1: Isocitrate/Isopropylmalate dehydrogenase-like [53659] (5 families) 
the constituent families form similar dimers
Family c.77.1.5: Phosphotransacetylase [102663] (2 proteins)
Pfam PF01515; contains extra beta-alpha unit between strands 2 and 3; closer relationships to the PdxA-like and PlsX-like families
Protein Ethanolamine utilization protein EutD [117727] (1 species) 
Species Escherichia coli [TaxId:562] [117728] (1 PDB entry) 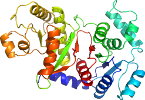
Domain d1vmia_: 1vmi A: [113678]
Structural genomics target
complexed with gol
Details for d1vmia_
PDB Entry: 1vmi (more details), 2.32 Å
PDB Description: Crystal structure of Putative phosphate acetyltransferase (np_416953.1) from Escherichia coli k12 at 2.32 A resolution
PDB Compounds: (A:) putative phosphate acetyltransferaseSCOP Domain Sequences for d1vmia_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1vmia_ c.77.1.5 (A:) Ethanolamine utilization protein EutD {Escherichia coli [TaxId: 562]}
tdpalraiiercrelalraparvvfpdaldqrvlkaaqylhqqglatpilvanpfelrqf
alshgvamdglqvidphgnlamreefahrwlaragektppdalekltdplmfaaamvsag
kadvciagnlsstanvlraglriiglqpgcktlssiflmlpqysgpalgfadcsvvpqpt
aaqladialasaetwraitgeeprvamlsfssngsarhpcvanvqqateivrerapklvv
dgelqfdaafvpevaaqkapasplqgkanvmvfpsleagnigykiaqrlggyravgpliq
glaapmhdlsrgcsvqeiielalvaavpr
SCOP Domain Coordinates for d1vmia_:
Click to download the PDB-style file with coordinates for d1vmia_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1vmia_:
- d1vmia_ first appeared in SCOP 1.71
- d1vmia_ appears in SCOP 1.75
- d1vmia_ appears in the current release, SCOPe 2.08, called d1vmia1

 View in 3D
View in 3D