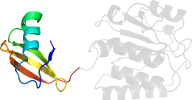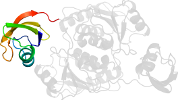Lineage for d1v6zb2 (1v6z B:67-228)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.116: alpha/beta knot [75216] (1 superfamily)
core: 3 layers: a/b/a, parallel beta-sheet of 5 strands, order 21435; contains a deep trefoil knot
Superfamily c.116.1: alpha/beta knot [75217] (9 families) 
known or predicted SAM-dependent methytransferases including the SPOUT 'sequence' superfamily
all known members have dimeric structures
Family c.116.1.5: YggJ C-terminal domain-like [89632] (4 proteins)
contains extra strand (3) in the parallel beta-sheet, order 321546; similar dimerisation to the MTH1 domain
Protein Hypothetical protein TTHA0657 (TT1575) [117494] (1 species) 
Species Thermus thermophilus [TaxId:274] [117495] (3 PDB entries)
Uniprot Q5SKI6
Domain d1v6zb2: 1v6z B:67-228 [113554]
Other proteins in same PDB: d1v6za1, d1v6zb1
Structural genomics target
Details for d1v6zb2
PDB Entry: 1v6z (more details), 2 Å
PDB Description: crystal structure of tt1573 from thermus thermophilus
PDB Compounds: (B:) hypothetical protein TTHA0657SCOPe Domain Sequences for d1v6zb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1v6zb2 c.116.1.5 (B:67-228) Hypothetical protein TTHA0657 (TT1575) {Thermus thermophilus [TaxId: 274]}
revgvevvlyvallkgdklaevvraatelgatriqplvtrhsvpkemgegklrrlraval
eaakqsgrvvvpevlppiplkavpqvaqglvahvgatarvrevldpekplalavgpeggf
aeeevalleargftpvslgrrilraetaalallalctagegr
SCOPe Domain Coordinates for d1v6zb2:
Click to download the PDB-style file with coordinates for d1v6zb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1v6zb2:
- d1v6zb2 first appeared in SCOP 1.71
- d1v6zb2 appears in SCOPe 2.06
- d1v6zb2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D