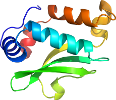
Lineage for d1uesd2 (1ues D:685-791)
- Root: SCOPe 2.05

Class d: Alpha and beta proteins (a+b) [53931] (381 folds) 
Fold d.44: Fe,Mn superoxide dismutase (SOD), C-terminal domain [54718] (1 superfamily)
alpha-beta(2)-alpha-beta-alpha(2); 3 strands of antiparallel sheet: 213
Superfamily d.44.1: Fe,Mn superoxide dismutase (SOD), C-terminal domain [54719] (2 families) 
automatically mapped to Pfam PF02777
Family d.44.1.1: Fe,Mn superoxide dismutase (SOD), C-terminal domain [54720] (4 proteins) 
Protein Cambialistic superoxide dismutase [54732] (2 species)
active with either fe or mn
Species Porphyromonas gingivalis [TaxId:837] [54734] (3 PDB entries)
Uniprot P19665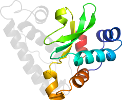
Domain d1uesd2: 1ues D:685-791 [107805]
Other proteins in same PDB: d1uesa1, d1uesb1, d1uesc1, d1uesd1
complexed with mn
Details for d1uesd2
PDB Entry: 1ues (more details), 1.6 Å
PDB Description: crystal structure of porphyromonas gingivalis sod
PDB Compounds: (D:) superoxide dismutaseSCOPe Domain Sequences for d1uesd2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1uesd2 d.44.1.1 (D:685-791) Cambialistic superoxide dismutase {Porphyromonas gingivalis [TaxId: 837]}
kggapkgklgeaidkqfgsfekfkeefntagttlfgsgwvwlasdangklsiekepnagn
pvrkglnplltfdvwehayyltyqnrradhlkdlwsivdwdivesry
SCOPe Domain Coordinates for d1uesd2:
Click to download the PDB-style file with coordinates for d1uesd2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1uesd2:
- d1uesd2 first appeared in SCOP 1.69
- d1uesd2 appears in SCOPe 2.04
- d1uesd2 appears in SCOPe 2.06
- d1uesd2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D