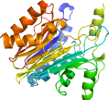
Lineage for d1t3m.2 (1t3m C:,D:)
- Root: SCOPe 2.06

Class d: Alpha and beta proteins (a+b) [53931] (385 folds) 
Fold d.153: Ntn hydrolase-like [56234] (2 superfamilies)
4 layers: alpha/beta/beta/alpha; has an unusual sheet-to-sheet packing
Superfamily d.153.1: N-terminal nucleophile aminohydrolases (Ntn hydrolases) [56235] (8 families) 
N-terminal residue provides two catalytic groups, nucleophile and proton donor
Family d.153.1.5: (Glycosyl)asparaginase [56261] (2 proteins)
automatically mapped to Pfam PF01112
Protein Glycosylasparaginase (aspartylglucosaminidase, AGA) [56262] (5 species)
the precursor chain is cleaved onto 2 fragments by autoproteolysis
Species Escherichia coli [TaxId:562] [103315] (3 PDB entries)
Uniprot P37595
isoaspartyl peptidase with L-asparaginase activity
putative L-asparaginase YbiK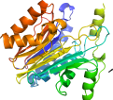
Domain d1t3m.2: 1t3m C:,D: [106362]
complexed with na, no3
Details for d1t3m.2
PDB Entry: 1t3m (more details), 1.65 Å
PDB Description: Structure of the isoaspartyl peptidase with L-asparaginase activity from E. coli
PDB Compounds: (C:) Putative L-asparaginase, (D:) Putative L-asparaginaseSCOPe Domain Sequences for d1t3m.2:
Sequence; same for both SEQRES and ATOM records: (download)
>g1t3m.2 d.153.1.5 (C:,D:) Glycosylasparaginase (aspartylglucosaminidase, AGA) {Escherichia coli [TaxId: 562]}
kaviaihggagaisraqmslqqelryiealsaivetgqkmleagesaldvvteavrllee
cplfnagigavftrdetheldacvmdgntlkagavagvshlrnpvlaarlvmeqsphvmm
igegaenfafargmervspeifstslryeqllaarkXtvgavaldldgnlaaatstggmt
nklpgrvgdsplvgagcyannasvavsctgtgevfiralaaydiaalmdygglslaeace
rvvmeklpalggsggliaidhegnvalpfntegmyrawgyagdtpttgiyr
SCOPe Domain Coordinates for d1t3m.2:
Click to download the PDB-style file with coordinates for d1t3m.2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1t3m.2:
- d1t3m.2 first appeared in SCOP 1.69
- d1t3m.2 appears in SCOPe 2.05
- d1t3m.2 appears in SCOPe 2.07
- d1t3m.2 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1t3m.1, d1t3m.1, d1t3m.1, d1t3m.1 |