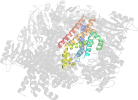
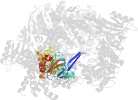
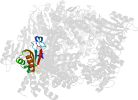Lineage for d3tuid1 (3tui D:1-240)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.37: P-loop containing nucleoside triphosphate hydrolases [52539] (1 superfamily)
3 layers: a/b/a, parallel or mixed beta-sheets of variable sizes
Superfamily c.37.1: P-loop containing nucleoside triphosphate hydrolases [52540] (27 families) 
division into families based on beta-sheet topologies
Family c.37.1.12: ABC transporter ATPase domain-like [52686] (25 proteins)
there are two additional subdomains inserted into the central core that has a RecA-like topology
missing some secondary structures that made up less than one-third of the common domain

Protein automated matches [190723] (8 species)
not a true protein
Species Escherichia coli K-12 [TaxId:83333] [233742] (1 PDB entry) 
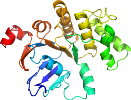
Domain d3tuid1: 3tui D:1-240 [233743]
Other proteins in same PDB: d3tuia_, d3tuib_, d3tuic2, d3tuic3, d3tuid2, d3tuie_, d3tuif_, d3tuig2, d3tuih2, d3tuih3
automated match to d3dhwc1
complexed with adp
Details for d3tuid1
PDB Entry: 3tui (more details), 2.9 Å
PDB Description: Inward facing conformations of the MetNI methionine ABC transporter: CY5 native crystal form
PDB Compounds: (D:) Methionine import ATP-binding protein metNSCOPe Domain Sequences for d3tuid1:
Sequence; same for both SEQRES and ATOM records: (download)
>d3tuid1 c.37.1.12 (D:1-240) automated matches {Escherichia coli K-12 [TaxId: 83333]}
miklsnitkvfhqgtrtiqalnnvslhvpagqiygvigasgagkstlircvnllerpteg
svlvdgqelttlseseltkarrqigmifqhfnllssrtvfgnvalpleldntpkdevkrr
vtellslvglgdkhdsypsnlsggqkqrvaiaralasnpkvllcdqatsaldpattrsil
ellkdinrrlgltillithemdvvkricdcvavisngelieqdtvsevfshpktplaqkf
SCOPe Domain Coordinates for d3tuid1:
Click to download the PDB-style file with coordinates for d3tuid1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3tuid1:
- d3tuid1 first appeared in SCOPe 2.03
- d3tuid1 appears in SCOPe 2.07
 View in 3D
View in 3D