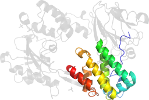
Lineage for d3sbtb2 (3sbt B:171-317)
- Root: SCOPe 2.08

Class a: All alpha proteins [46456] (290 folds) 
Fold a.118: alpha-alpha superhelix [48370] (28 superfamilies)
multihelical; 2 (curved) layers: alpha/alpha; right-handed superhelix
Superfamily a.118.29: U5 small nuclear riboprotein particle assembly factor Aar2 C-terminal domain-like [345916] (1 family) 
C-terminal part of Pfam PF05282
Structural similarity with VHS domains (48464) noted in PubMed 21764848, but no compelling evidence of homology
Family a.118.29.1: U5 small nuclear riboprotein particle assembly factor Aar2 C-terminal domain-like [345957] (2 proteins) 
Protein U5 small nuclear riboprotein particle assembly factor Aar2 C-terminal domain [346042] (1 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:559292] [346205] (158 PDB entries) 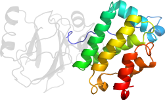
Domain d3sbtb2: 3sbt B:171-317 [343864]
Other proteins in same PDB: d3sbta1, d3sbta2, d3sbtb1
automated match to d3sbsa2
complexed with pgo, so4
Details for d3sbtb2
PDB Entry: 3sbt (more details), 1.8 Å
PDB Description: Crystal structure of a Aar2-Prp8 complex
PDB Compounds: (B:) A1 cistron-splicing factor AAR2SCOPe Domain Sequences for d3sbtb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d3sbtb2 a.118.29.1 (B:171-317) U5 small nuclear riboprotein particle assembly factor Aar2 C-terminal domain {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 559292]}
dpahslnytvinfksreairpghemedfldksyylntvmlqgifknssnyfgelqfafln
amffgnygsslqwhamielicssatvpkhmldkldeilyyqiktlpeqysdillnervwn
iclyssfqknslhntekimenkypell
SCOPe Domain Coordinates for d3sbtb2:
Click to download the PDB-style file with coordinates for d3sbtb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3sbtb2:
- d3sbtb2 first appeared in SCOPe 2.07

 View in 3D
View in 3D