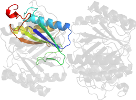
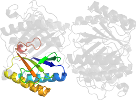
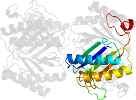
Lineage for d2aaga1 (2aag A:1-129)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.80: Tautomerase/MIF [55330] (1 superfamily)
(beta-alpha-beta)2; 2 layers: alpha/beta; mixed beta-sheet
generally forms trimers with three closely packed beta-sheets
Superfamily d.80.1: Tautomerase/MIF [55331] (6 families) 

Family d.80.1.6: MSAD-like [143532] (1 protein) 
Protein Malonate semialdehyde decarboxylase, MSAD [143533] (1 species) 
Species Pseudomonas pavonaceae [TaxId:47881] [143534] (3 PDB entries)
Uniprot Q9EV83 2-130! Uniprot Q9EV83 2-230
Domain d2aaga1: 2aag A:1-129 [126475]
automatically matched to 2AAL A:1-129
mutant
Details for d2aaga1
PDB Entry: 2aag (more details), 1.85 Å
PDB Description: crystal structures of the wild-type, mutant-p1a and inactivated malonate semialdehyde decarboxylase: a structural basis for the decarboxylase and hydratase activities
PDB Compounds: (A:) Malonate Semialdehyde DecarboxylaseSCOP Domain Sequences for d2aaga1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2aaga1 d.80.1.6 (A:1-129) Malonate semialdehyde decarboxylase, MSAD {Pseudomonas pavonaceae [TaxId: 47881]}
pllkfdlfygrtdaqikslldaahgamvdafgvpandryqtvsqhrpgemvledtglgyg
rssavvlltvisrprseeqkvcfyklltgalerdcgispddvivalvensdadwsfgrgr
aefltgdlv
SCOP Domain Coordinates for d2aaga1:
Click to download the PDB-style file with coordinates for d2aaga1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2aaga1:
- d2aaga1 first appeared in SCOP 1.73
- d2aaga1 is called d2aaga_ in SCOPe 2.01
- d2aaga1 appears in the current release, SCOPe 2.08, called d2aaga_
 View in 3D
View in 3D