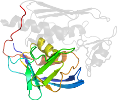
Lineage for d1xxea2 (1xxe A:128-270)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.14: Ribosomal protein S5 domain 2-like [54210] (1 superfamily)
core: beta(3)-alpha-beta-alpha; 2 layers: alpha/beta; left-handed crossover
Superfamily d.14.1: Ribosomal protein S5 domain 2-like [54211] (12 families) 
Family d.14.1.7: UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC [89827] (1 protein)
duplication; there are two structural repeats of this fold; each repeat is elaborated with additional structures forming the active site
Protein UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC [89828] (1 species) 
Species Aquifex aeolicus [TaxId:63363] [89829] (11 PDB entries)
Uniprot O67648 3-270
Domain d1xxea2: 1xxe A:128-270 [116158]
complexed with tux, zn
Details for d1xxea2
PDB Entry: 1xxe (more details)
PDB Description: rdc refined solution structure of the aalpxc/tu-514 complex
PDB Compounds: (A:) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylaseSCOP Domain Sequences for d1xxea2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1xxea2 d.14.1.7 (A:128-270) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC {Aquifex aeolicus [TaxId: 63363]}
epiivedegrlikaepsdtlevtyegefknflgrqkftfvegneeeivlartfcfdweie
hikkvglgkggslkntlvlgkdkvynpeglryenepvrhkvfdligdlyllgspvkgkfy
sfrgghslnvklvkelakkqklt
SCOP Domain Coordinates for d1xxea2:
Click to download the PDB-style file with coordinates for d1xxea2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1xxea2:
- d1xxea2 first appeared in SCOP 1.71
- d1xxea2 appears in SCOP 1.73
- d1xxea2 appears in SCOPe 2.01
- d1xxea2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D